Projects
Three themes dominate research in the Harris Laboratory:
Viral Restriction, Cancer Mutagenesis, and Base-Editing Technology
I. DNA Deamination in Retrovirus and Transposon Restriction
APOBEC3 proteins are potent inhibitors of many different types of retroviruses and retrotransposons (collectively retroelements). A prototype for understanding this host-pathogen conflict is the mechanism of HIV-1 restriction by APOBEC3D, F, G, and H (see figure below). These APOBEC3 proteins enter assembling viral particles, travel with the particle until a new target cell is infected, and then deaminate viral cDNA cytosines to uracils during reverse transcription. These lesions often manifest as viral G-to-A hypermutations. Many projects in the Harris Laboratory are dedicated to better understanding this restriction mechanism and how HIV-1 uses its accessory protein Vif to escape restriction.
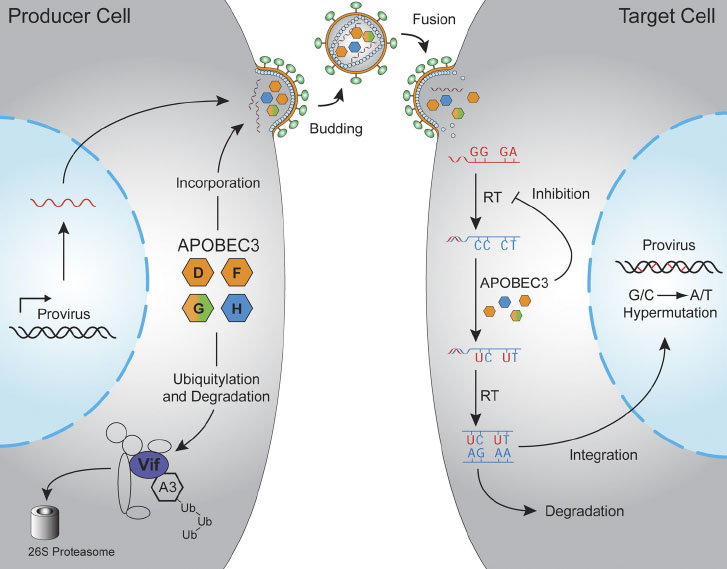
Hultquist et. al. (2011). Journal of Virology, 85, 11220-34.
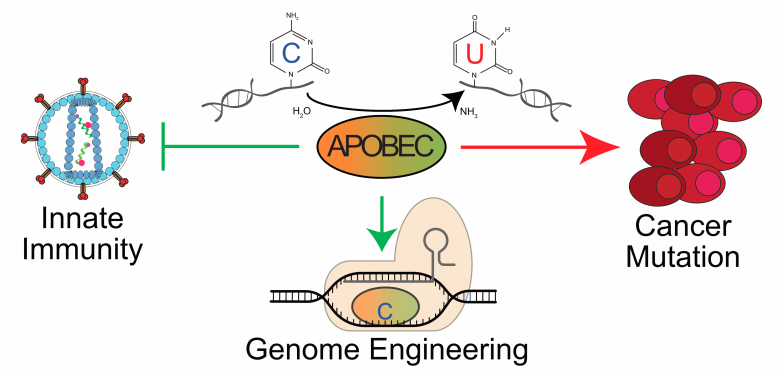
II. DNA Deamination in Cancer
The expression of enzymes that catalyze DNA C-to-U deamination poses an inherent threat to cells. Undoubtedly, many mechanisms exist to minimize these threats and maximize beneficial outcomes. Nevertheless, considerable evidence supports the hypothesis that enzyme-catalyzed DNA deamination contributes to many distinct stages of cancer progression, from the initial stages of transformation to the evolution of drug resistance (see figure below). Several projects in the Harris Laboratory are dedicated to understanding how DNA deaminases contribute to human carcinogenesis and developing novel methods to mitigate this pathogenic outcome.
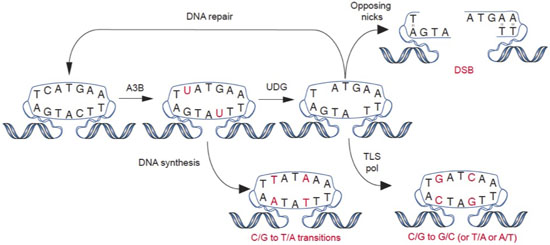
Adapted from Refsland & Harris. (2013). Current Topics in Microbiology and Immunology, 371, 1-27.
III. APOBEC-Cas9 Base-Editing Technology
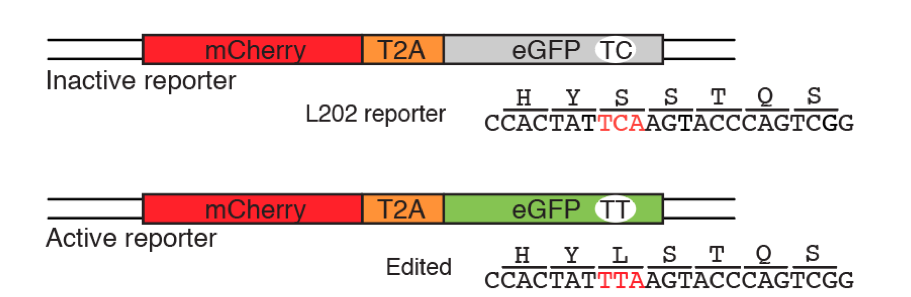
More recently, APOBEC enzymes have been implemented as precise base-editing tools in the field of genome engineering. With the advent of CRISPR-Cas9 technology, and the sequence specificity these systems permit, researchers have tethered deaminase enzymes to Cas9 nickase proteins to generate specific C-to-T changes at discrete nucleotide locations. Furthermore, the Harris lab has developed fluorescence-based reporter assays to test APOBEC-Cas9 base-editing systems (St. Martin et al., 2018, Nucleic Acid Res.), giving rise to tractable cell systems to optimize current deaminase editors, test novel editing tools, and perform large scale screens for editing activity in vitro (see figure below). This work has widespread implications in the genome-editing field, paving the way for future genetic disease correction technologies and therapeutics.
I am text block. Click edit button to change this text. Lorem ipsum dolor sit amet, consectetur adipiscing elit. Ut elit tellus, luctus nec ullamcorper mattis, pulvinar dapibus leo.