Projects background
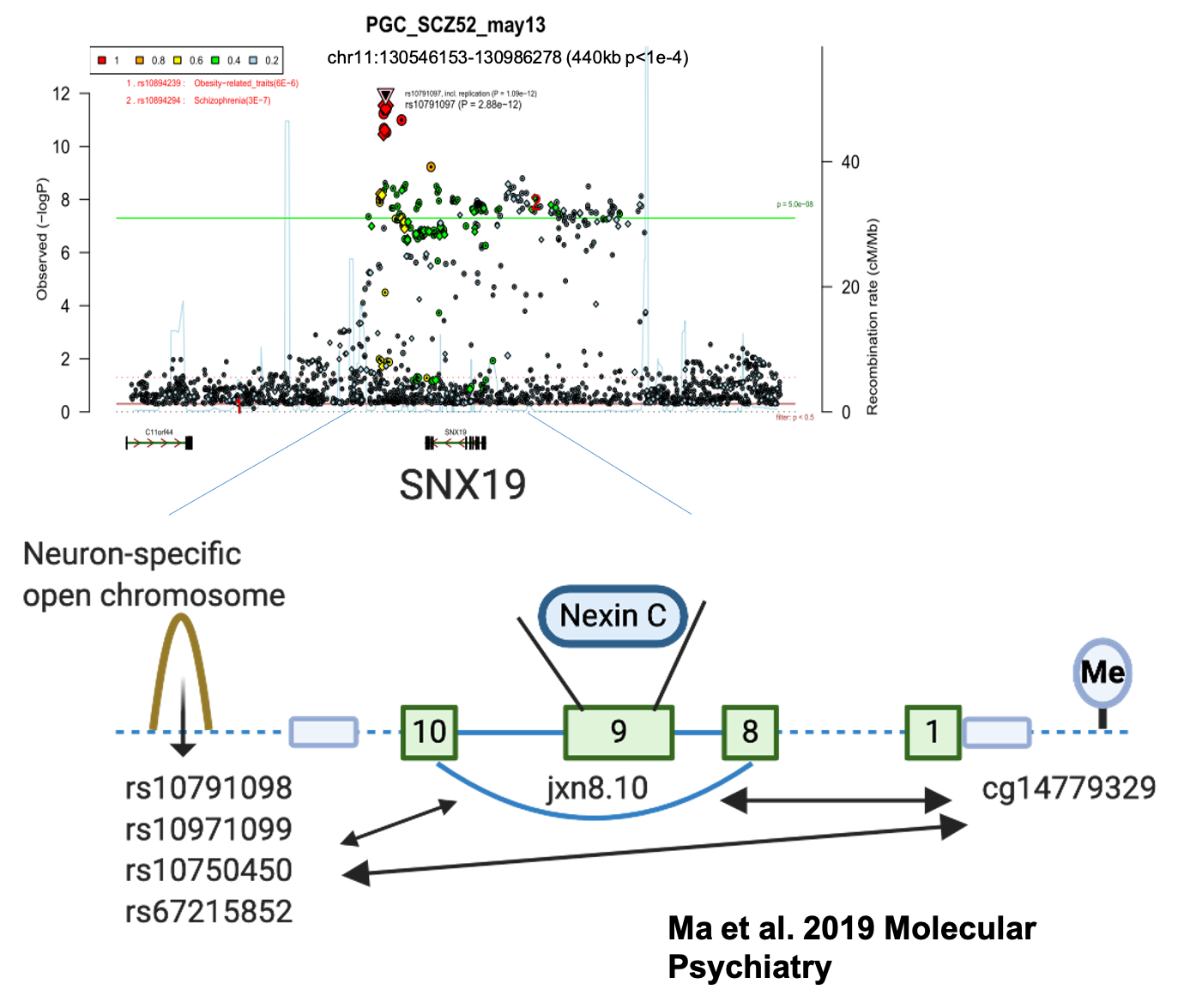
Based on schizophrenia GWAS data, we discovered a specific cluster of SNX19 gene transcripts associated with schizophrenia risk using RNA-seq and DNA methylation data combined with the genotyping data generated from 495 postmortem brains of schizophrenia and controls. We also investigated SNX19 transcript patterns in autopsy brains through molecular and cell biology experiments (Ma et al. 2020).
We are using CRISPR gene editing on human induced pluripotent stem cells (hiPSCs) to create isogenic lines, followed by differentiating them into 2D neurons and 3D brain organoids to study the impact of SNX19 on neuronal function (e.g., synapse loss, calcium level, and autophagy). We are performing a series of behavior tests in mouse model to investigate the impact of SNX19 on cognitive functions.
This work is funded by NIH and the Alzheimer’s Association.
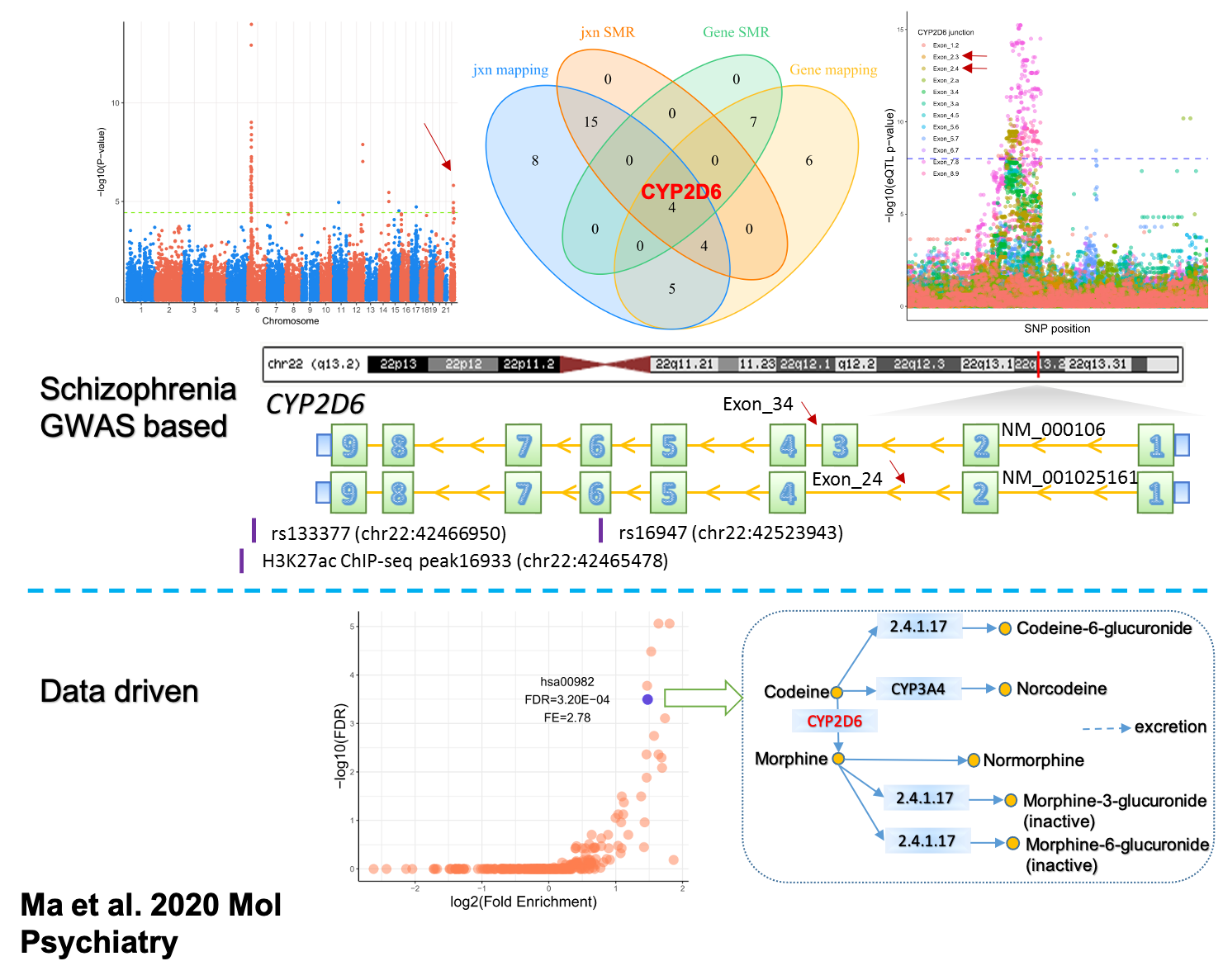
I extended the investigation of functional genomic regions of schizophrenia from one gene to the whole genome to analyze RNA-seq data from 1,479 human postmortem brains combined with their whole-genome sequencing data. While confirming SNX19 splicing events in schizophrenia risk, I also identified alternative transcripts of CYP2D6 predisposed to schizophrenia. In parallel, I determined causal variations of CYP2D6 transcription by ChIP-seq and DNA methylation data generated from another cohort of human postmortem brains. Finally, convergence upon CYP2D6 as an essential candidate gene was identified again using a data-driven enrichment analysis (Ma et al. 2021). We are investigating the role of CYP2D6 in brain using the mouse model and human iPSC-derived models.
Project - APOE in Alzheimer's disease
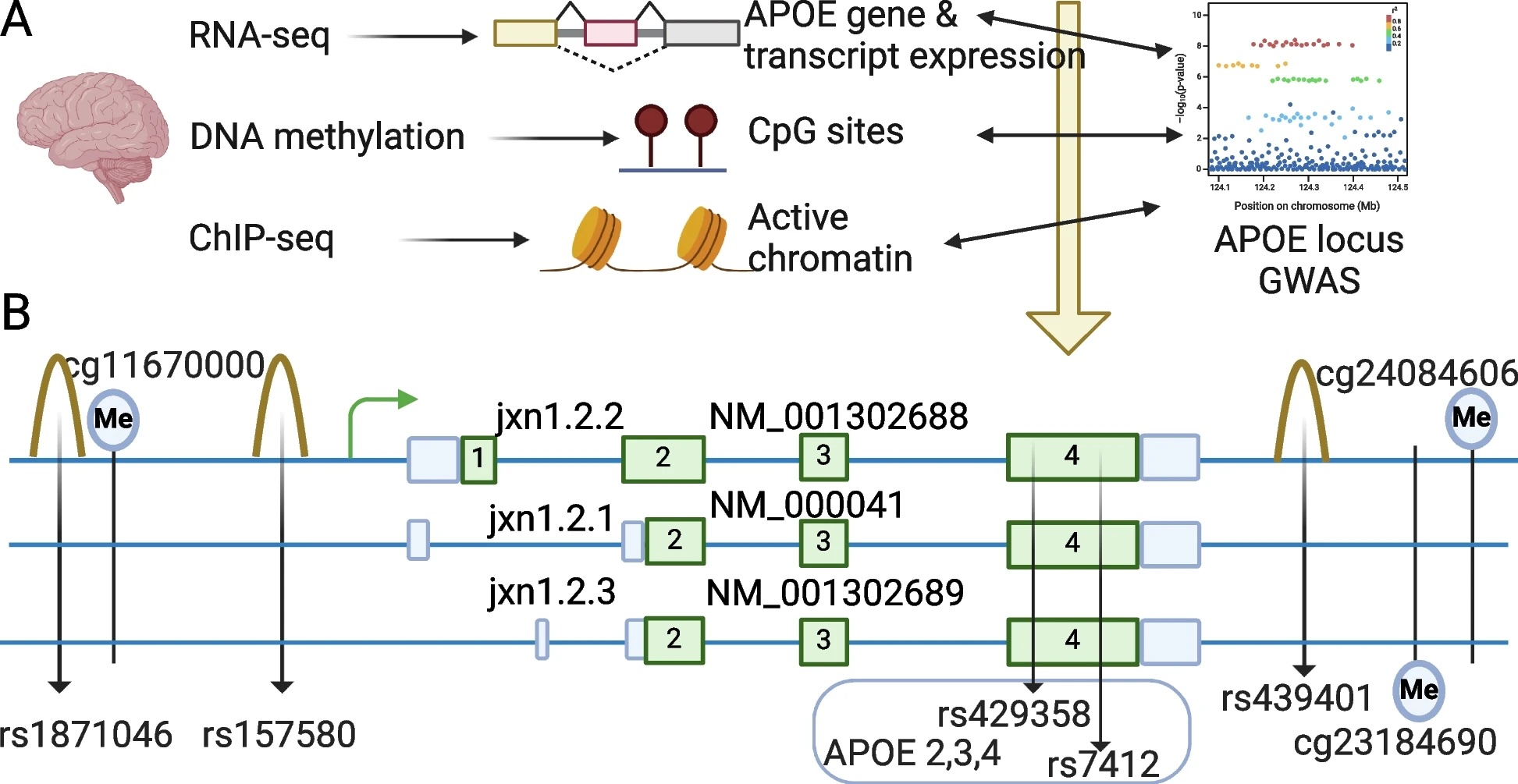
APOE4 is the strongest genetic risk factor in Alzheimer’s disease. However, just having APOE4 doesn’t mean someone will definitely develop Alzheimer’s, and not having it doesn’t eliminate the risk either.
Recent large-scale genetic studies have uncovered additional variations in the APOE region. These variations have been linked to Alzheimer’s risk, but the challenge has been figuring out which APOE mRNA these variations affect in the brain.
To explore this further, we analyzed brain data from deceased individuals. For the first time, we discovered a specific version of the APOE gene, called the jxn1.2.2 transcript, that may increase Alzheimer’s risk in people of European and African ancestry, regardless of their APOE2, APOE3, or APOE4 type.
This work has been published on Molecular Neurodegeneration and is supported by the Alzheimer’s Association and the National Alzheimer’s Coordinating Center New Investigator Award (NIAP). We are leveraging CRISPR genome editing using human iPSC-derived models to study the risk factors we identified.
Project - Cell2Spatial

Cell2Spatial: Reconstructing Tissue Architecture at Single-Cell Granularity to map single-cell data onto spatial transcriptomic spots (Li et al. 2025 PLOS Biology).
Ongoing Project
Genome-wide study
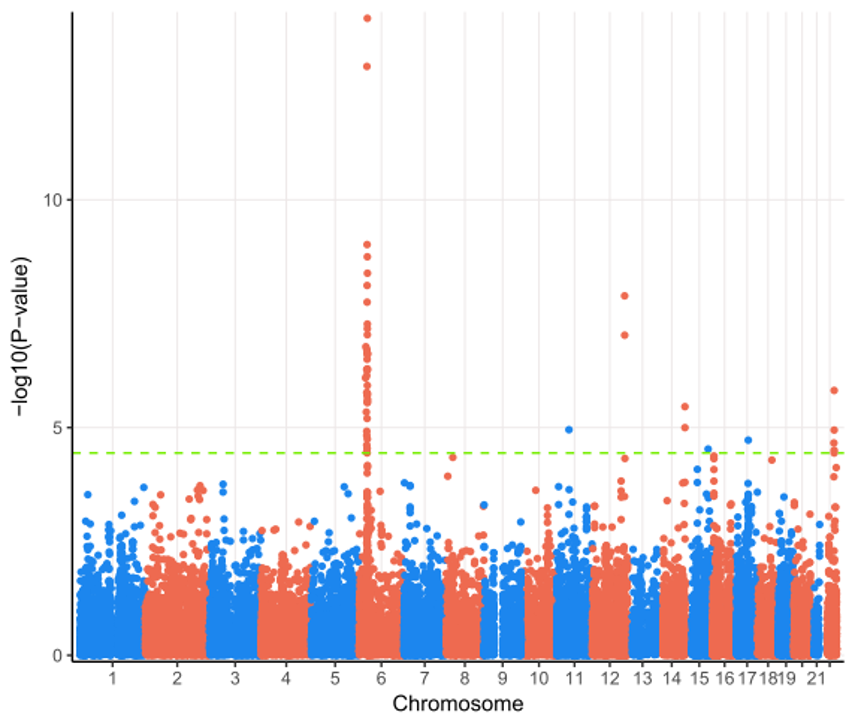
We perform genome-wide association between genetic variants and gene expression features, DNA methylations, histone markers, and more.
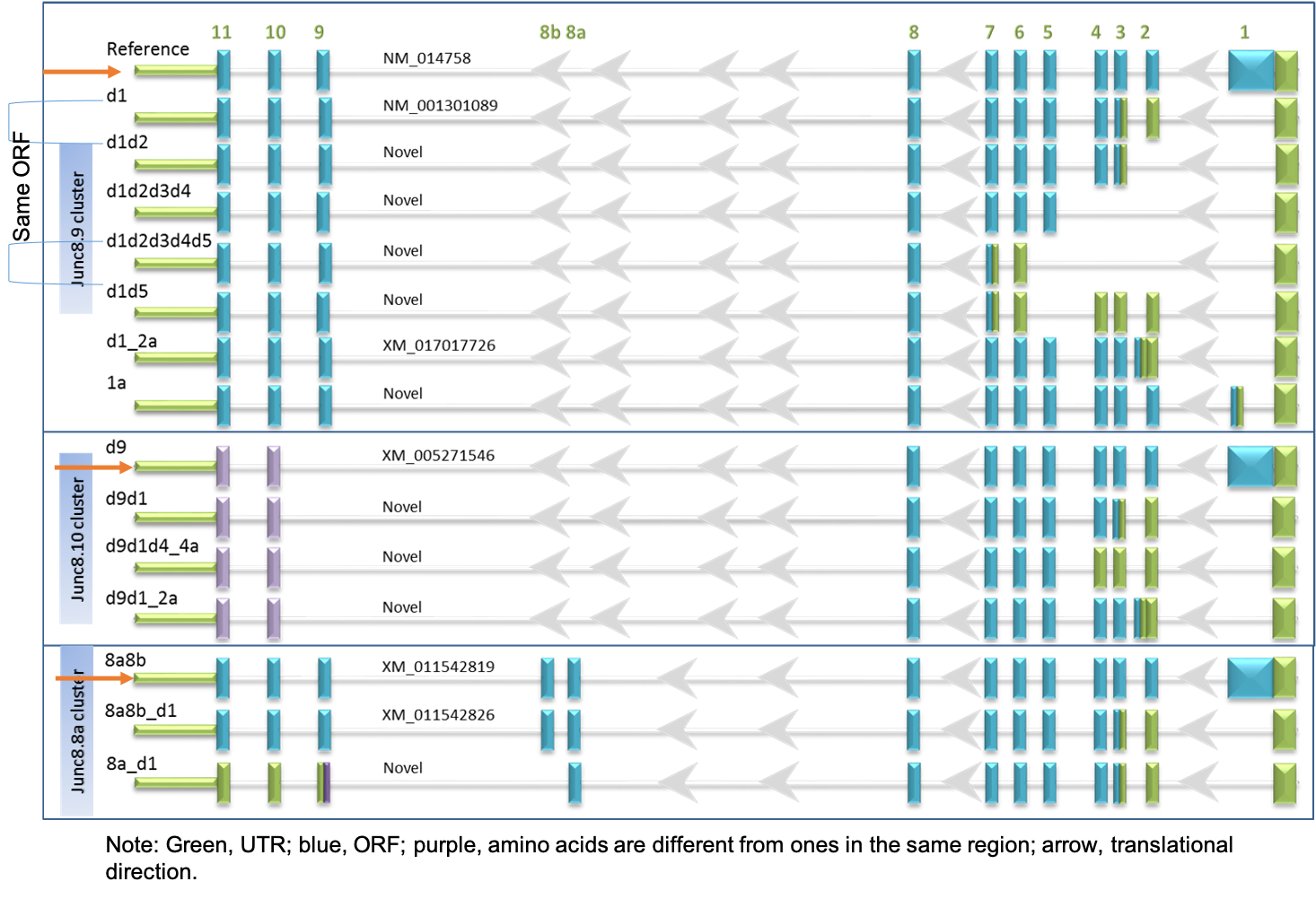
Transcripts diversity
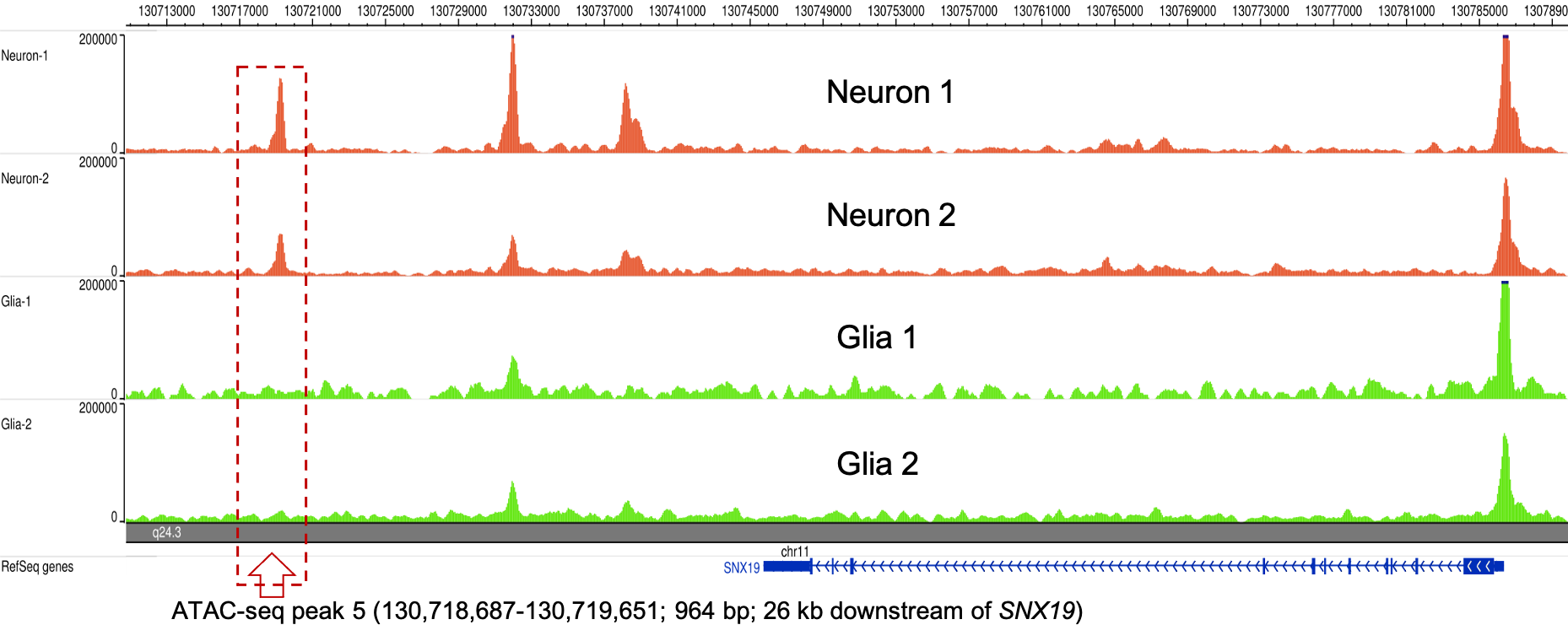
Epigenetic markers
We are combing short-read RNA-seq and long-read sequencing to decipher transcript expression patterns in human brains.
We use DNA methylation, ChIP-seq, and ATAC-seq to study the gene regulatory mechanisms.
CRISPR gene editing
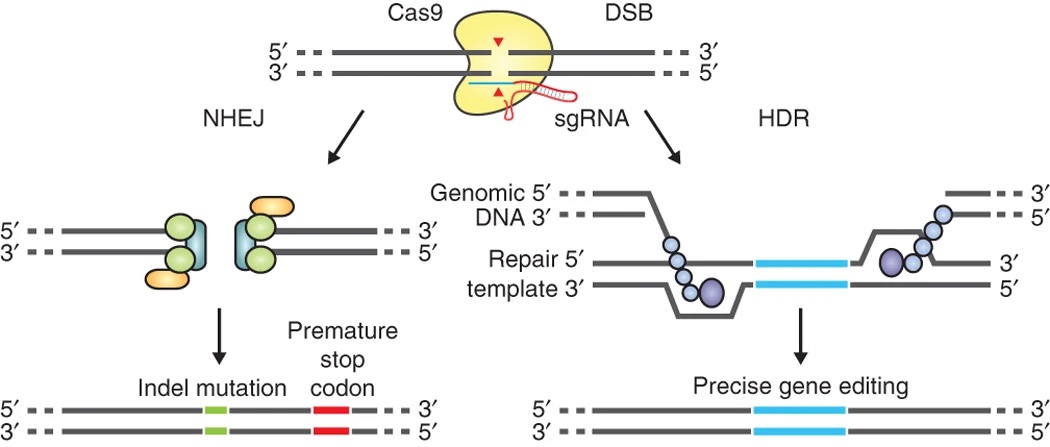
We are using CRISPR editing technology (Ran et al. 2013 Nat Protoc) on the disease genes we prioritized (e.g., SNX19).
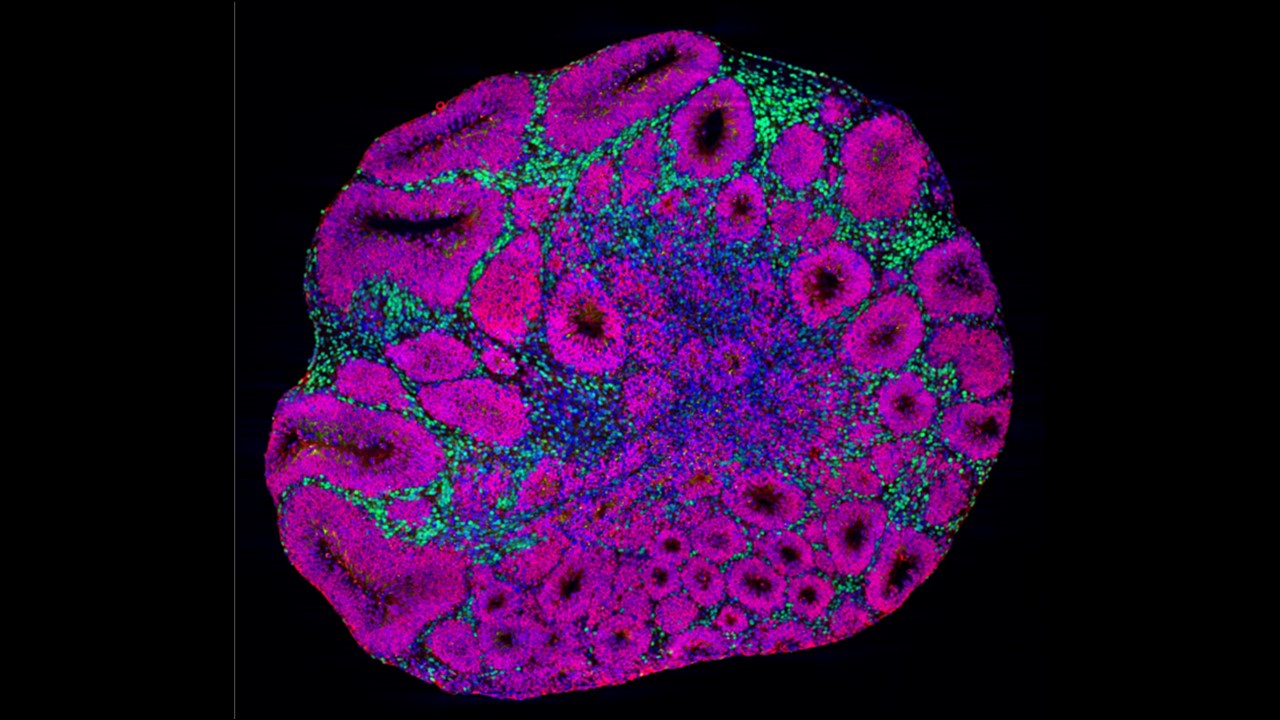
Figure legend:
A brain organoid showing neuron precursors (magenta) and deep-layer projection neurons (green).